Building extensions between Python libraries for Biotechnology laboratories

Hello again! This is Luiza, a GSoC contributor for the LabOp Project. My task is to build bridges between programming languages for Biotechnology Laboratory automation.
When talking about life sciences, reproducibility is a issue amongst most research centers. Biotechnology focused laboratories usually have their own protocols developed in house for their own applications. Researchers rely on such protocols to perform their experiments and collect data but when it comes to sharing those protocols and performing them in different laboratories many difficulties arise. Whether it is by lack of equipment, reagents or even by having different orders of execution, replicating a protocol in another laboratory is a challenge. To address this issue LabOp was developed to represent a protocol and convert it in many ways possible, so it can be executed by humans and by machines.
PylabRobot and PyHamilton also come to the picture as such libraries exist to make it possible to write protocols for Hamilton robots(and Tecan machines as well for PylabRobot) but those libraries share the limitation of being able to only represent laboratory protocols at their lower levels, with the user having to write every single command in Python for the protocol to be executed. Thus I’m currently developing an extension for LabOp protocols to be converted into PylabRobot/PyHamilton scripts. This way the researcher writing the protocol can do it in a friendlier fashion, using human-friendly terms to write protocols for robot execution.
The first step is building a correspondence spreadsheet with a hello world protocol written in both languages (LabOp | PylabRobot ). This way we can make an equivalence between the functions, parameters and default commands of both Libraries, as well as their structure. This spreadsheet will serve as guidance for the conversion of the Liquid handling steps from their representation in LabOp to their representation in Pylabrobot.
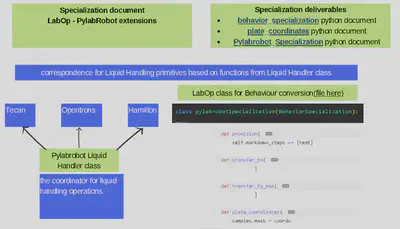
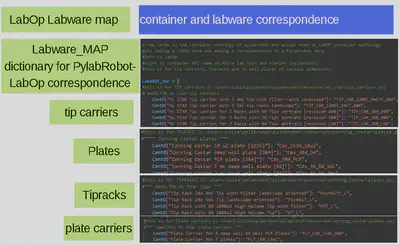
The second step is to create a file that’ll do execute the conversion. In this file I will define a Labware map that’s basically a dictionary translating the resources LabOp names into Labware IDs recognizable by PylabRobots “resource” classes and a Behaviourspecialization class that should convert LabOp actions into PylabRobots Liquid Handler class operations as they’ll coordinate the commands sent from the script to the machines.(see featured images)
Then we move to the protocol that will be tested on the Hamilton Machines, this is a Plasmid purification protocol that is usually performed by a human at a very lower level, one sample at a time. This limitation is not present on Hamilton robots as they can handle many samples at the same time with only one protocol execution. The robot that will be running this protocol has two modules that are not yet present in PylabRobot’s extensions, a pressure pump module and a on deck heatershaker. I’ll be implemmenting this modules in PylabRobot based on their default commands present in PyHamilton and run the protocol on a Hamilton Starlet unit.
The steps of the protocol have been decoupled to facilitate the pilot testing, they are as follows:
- Liquid handling - GOOD TO GO
- Pressure pump module- requires adjustments
- plate grippers(necessary to move the plasmid plate from one module to another) - requires adjustment
- On deck heaterShaker- GOOD TO GO
The first pilot tests of the protocol will be run with water instead of plasmid to verify that all the steps are going smoothly, when that’s out of the way we will perform the protocol with dirty plasmids that require purification (which is what the protocol is for). The measurements for success will be sequencing the plasmid (if possible), performing a gel eletrophoresis and measuring absorbance of the DNA.
The goal of this tests is to gather data from the efectiveness of the protocol and its execution on the machine, thus confirming that it is in fact a useful mechanism for DNA purification.